Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure.
N Engl J Med. 2020
"A European collaborative study with participation of Hospital Ramón y Cajal identifies genetic susceptibility in patients with COVID-19 infection and respiratory failure" – Dr. Agustín Albillos –
Summary:
BACKGROUND: There is considerable variation in disease behavior among patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (Covid-19). Genomewide association analysis may allow for the identification of potential genetic factors involved in the development of Covid-19.
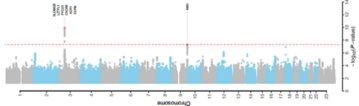
METHODS: We conducted a genomewide association study involving 1980 patients with Covid-19 and severe disease (defined as respiratory failure) at seven hospitals in the Italian and Spanish epicenters of the SARS-CoV-2 pandemic in Europe. After quality control and the exclusion of population outliers, 835 patients and 1255 control participants from Italy and 775 patients and 950 control participants from Spain were included in the final analysis. In total, we analyzed 8,582,968 single-nucleotide polymorphisms and conducted a meta-analysis of the two case-control panels.
RESULTS: We detected cross-replicating associations with rs11385942 at locus 3p21.31 and with rs657152 at locus 9q34.2, which were significant at the genomewide level (P<5×10−8) in the meta-analysis of the two case-control panels (odds ratio, 1.77; 95% confidence interval [CI], 1.48 to 2.11; P = 1.15×10−10; and odds ratio, 1.32; 95% CI, 1.20 to 1.47; P = 4.95×10−8, respectively). At locus 3p21.31, the association signal spanned the genes SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6 and XCR1. The association signal at locus 9q34.2 coincided with the ABO blood group locus; in this cohort, a blood-group-specific analysis showed a higher risk in blood group A than in other blood groups (odds ratio, 1.45; 95% CI, 1.20 to 1.75; P = 1.48×10−4) and a protective effect in blood group O as compared with other blood groups (odds ratio, 0.65; 95% CI, 0.53 to 0.79; P = 1.06×10−5).
CONCLUSIONS: We identified a 3p21.31 gene cluster as a genetic susceptibility locus in patients with Covid-19 with respiratory failure and confirmed a potential involvement of the ABO blood-group system. (Funded by Stein Erik Hagen and others.)
Why do you highligth this publication?
· Variants from two regions of the human genome are associated with an increased risk of respiratory failure in patients infected with coronavirus.
· One of the variants is located on chromosome 3 (rs11385942) and can affect the expression of genes that help the entry of the virus and the development of cytokine storm, by being related to the inflammatory immune response in the lungs against pathogens. This is more frequent in people with an average of 59 years, which would partially explain the severity of certain cases in this age group.
· The second region is located on chromosome 9 (rs657152), in the gene that determines the blood group of the ABO system. Seeing that blood group A is associated with 50% more vulnerability to the development of respiratory failure, while group O is protective.
· The frequency of both variants on chromosomes 3 and 9 is significantly higher in patients who required mechanical ventilation compared to those who were administered oxygen, regardless of age and sex.
Publication commented by:
Dr. Agustín Albillos
Departament of Gastroenterology
Liver and Digestive Diseases Group-IRYCIS
University of Alcalá